Membrane Bound Collection
"Exploring the Intricate World of Membrane-Bound Cell Structures" The Golgi apparatus, as seen through a scanning electron microscope (SEM
All Professionally Made to Order for Quick Shipping
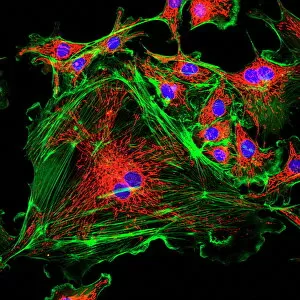
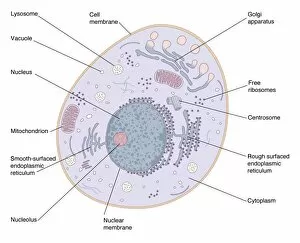
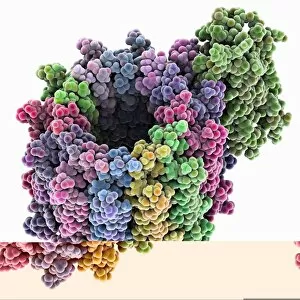
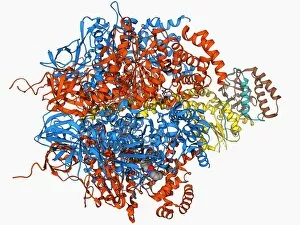
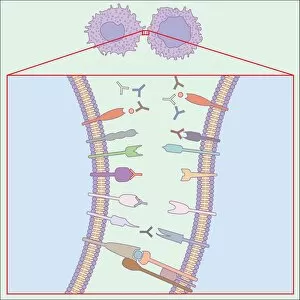
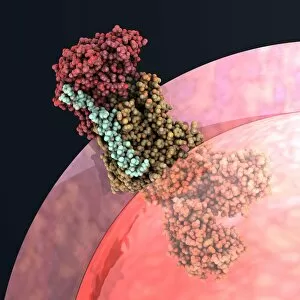
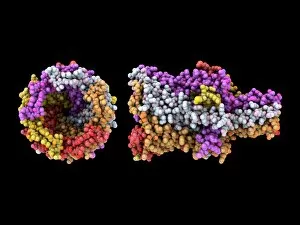
"Exploring the Intricate World of Membrane-Bound Cell Structures" The Golgi apparatus, as seen through a scanning electron microscope (SEM), reveals its fascinating membrane-bound structure. This organelle plays a crucial role in modifying, sorting, and packaging proteins for transport within the cell. Another captivating view under SEM showcases the mitochondria, known as the powerhouse of the cell. These membrane-bound structures generate energy by converting nutrients into ATP molecules. The intricate details captured by SEM highlight their importance in cellular respiration. Delving deeper into animal cell structure, we observe another stunning SEM image showcasing mitochondria at work. These dynamic organelles are responsible for producing ATP through oxidative phosphorylation and play an essential role in various cellular processes. Zooming even closer to molecular level, we encounter remarkable images of ATP synthase molecule C014 / 0880 and ATPase molecule F006 / 9300. These enzymes are integral to energy production within cells and facilitate the conversion between ADP and ATP. Further exploration leads us to witness a molecular model depicting both ATPase and inhibitor F006 / 9448. This visual representation sheds light on how inhibitors can regulate or inhibit this vital enzyme's activity, offering insights into potential therapeutic interventions. Intriguingly, another image captures an isolated view of an individual ATPase molecule F006 / 9258. Its complex structure highlights its significance in hydrolyzing ATP to release energy required for various cellular functions. Diving deeper into specific transport mechanisms across membranes, we encounter a striking depiction of sucrose-specific porin molecule F006 / 9218. This protein allows selective passage of sucrose molecules across membranes while maintaining cellular homeostasis. Overall, these captivating images provide glimpses into the mesmerizing world of membrane-bound structures within cells – from macroscopic organelles like Golgi apparatus and mitochondria observed through SEM to individual molecules such as ATP synthase and ATPase.